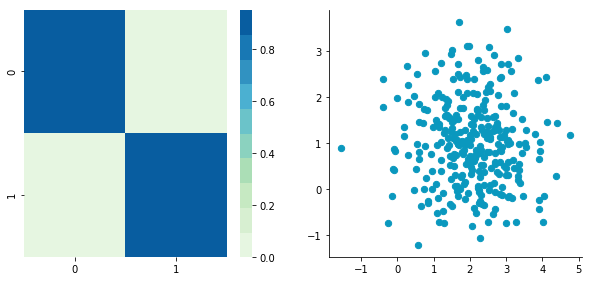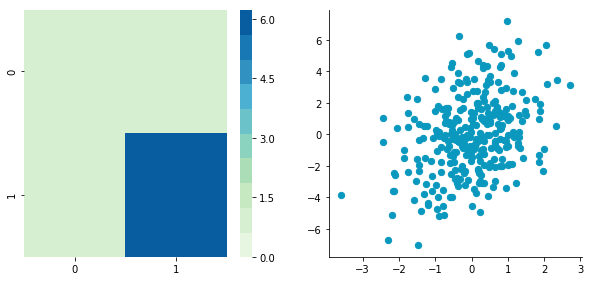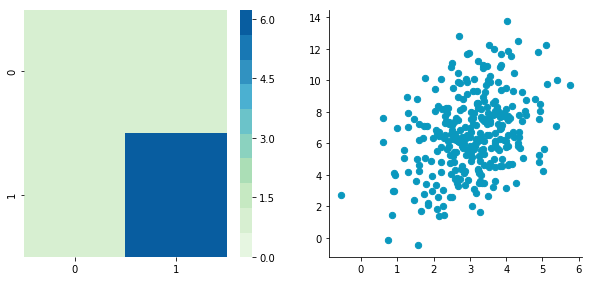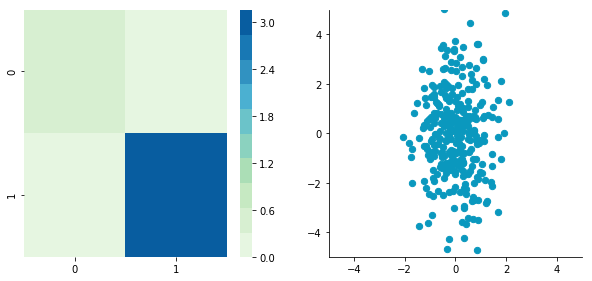что такое data whitening
Preprocessing for deep learning: from covariance matrix to image whitening
Last update: Jan. 2021
A notebook version of this post can be found here on Github.
The goal of this post/notebook is to go from the basics of data preprocessing to modern techniques used in deep learning. My point is that we can use code (Python/Numpy etc.) to better understand abstract mathematical notions! Thinking by coding! 💥
We will start with basic but very useful concepts in data science and machine learning/deep learning like variance and covariance matrix and we will go further to some preprocessing techniques used to feed images into neural networks. We will try to get more concrete insights using code to actually see what each equation is doing!
We call preprocessing all transformations on the raw data before it is fed to the machine learning or deep learning algorithm. For instance, training a convolutional neural network on raw images will probably lead to bad classification performances (Pal & Sudeep, 2016). The preprocessing is also important to speed up training (for instance, centering and scaling techniques, see Lecun et al., 2012; see 4.3).
Here is the syllabus of this tutorial:
Background: In the first part, we will get some reminders about variance and covariance and see how to generate and plot fake data to get a better understanding of these concepts.
Preprocessing: In the second part, we will see the basics of some preprocessing techniques that can be applied to any kind of data: mean normalization, standardisation and whitening.
Whitening images: In the third part, we will use the tools and concepts gained in 1. and 2. to do a special kind of whitening called Zero Component Analysis (ZCA). It can be used to preprocess images for deep learning. This part will be very practical and fun ☃️!
Feel free to fork the notebook. For instance, check the shapes of the matrices each time you have a doubt 🙂
1. Background
A. Variance and covariance
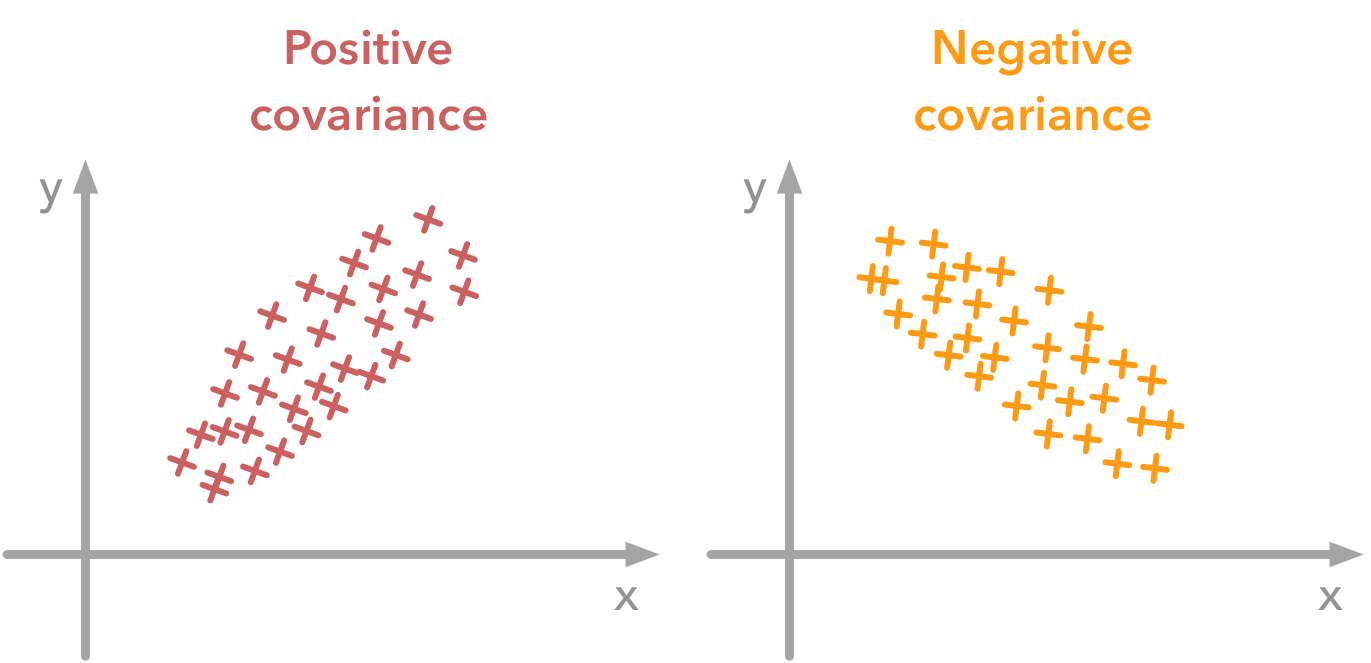
The variance of a variable describes how much the values are spread. The covariance is a measure that tells the amount of dependency between two variables. A positive covariance means that values of the first variable are large when values of the second variables are also large. A negative covariance means the opposite: large values from one variable are associated with small values of the other. The covariance value depends on the scale of the variable so it is hard to analyse it. It is possible to use the correlation coefficient that is easier to interpret. It is just the covariance normalized.
The covariance matrix is a matrix that summarizes the variances and covariances of a set of vectors and it can tell a lot of things about your variables. The diagonal corresponds to the variance of each vector:
Let’s just check with the formula of the variance:
Covariance
Example 1.
We will calculate the covariance between the first and the third column vectors:
Ok, great! That the value of the covariance matrix.
Let’s create the array first:
Now we will calculate the covariance with the Numpy function:
Finding the covariance matrix with the dot product
We end up with the same result as before!
The explanation is simple. The dot product between two vectors can be expressed:
That’s right, it is the sum of the products of each element of the vectors:
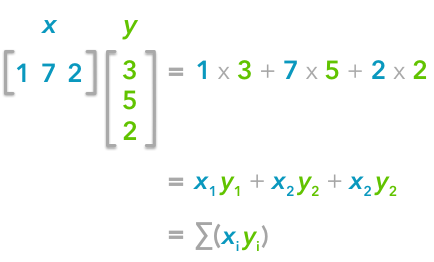
You can note that this is not too far from the formula of the covariance we have seen above:
The only difference is that in the covariance formula we subtract the mean of a vector to each of its elements. This is why we need to center the data before doing the dot product.
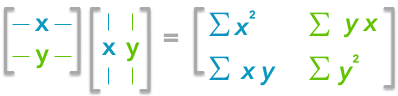
This is the covariance matrix! 🌵
B. Visualize data and covariance matrices
In order to get more insights about the covariance matrix and how it can be useful, we will create a function used to visualize it along with 2D data. You will be able to see the link between the covariance matrix and the data.
This function will calculate the covariance matrix as we have seen above. It will create two subplots: one for the covariance matrix and one for the data. The heatmap function from Seaborn is used to create gradients of color: small values will be colored in light green and large values in dark blue. The data is represented as a scatterplot. We choose one of our palette colors, but you may prefer other colors 🌈.
C. Simulating data
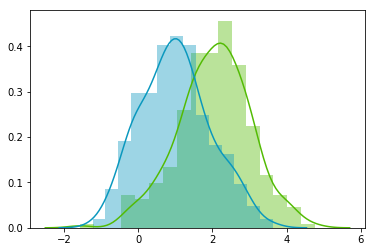
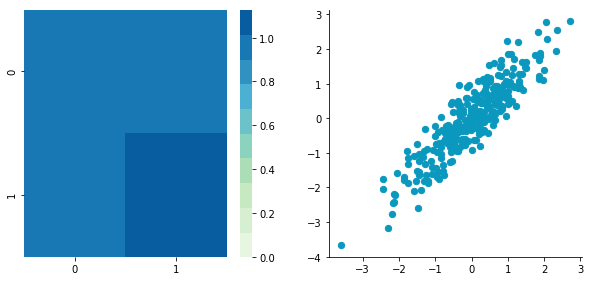
Uncorrelated data

This function needs the mean, the standard deviation and the number of observations of the distribution as input. We will create two random variables of 300 observations with a standard deviation of 1. The first will have a mean of 1 and the second a mean of 2. If we draw two times 300 observations from a normal distribution, both vectors will be uncorrelated.
Note 2: We use np.random.seed function for reproducibility. The same random number will be used the next time we run the cell!
Let’s check how the data looks like:
Nice, we have our two columns vectors.
Now, we can check that the distributions are normal:
Looks good! We can see that the distributions have equivalent standard deviations but different means (1 and 2). So that’s exactly what we have asked for!
Now we can plot our dataset and its covariance matrix with our function:
We can see on the scatterplot that the two dimensions are uncorrelated. Note that we have one dimension with a mean of 1 and the other with the mean of 2. Also, the covariance matrix shows that the variance of each variable is very large (around 1) and the covariance of columns 1 and 2 is very small (around 0). Since we insured that the two vectors are independent this is coherent (the opposite is not necessarily true: a covariance of 0 doesn’t guaranty independency (see here).
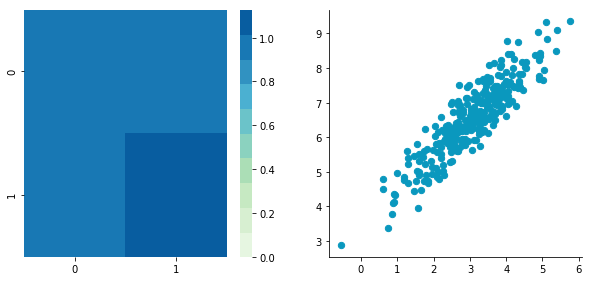
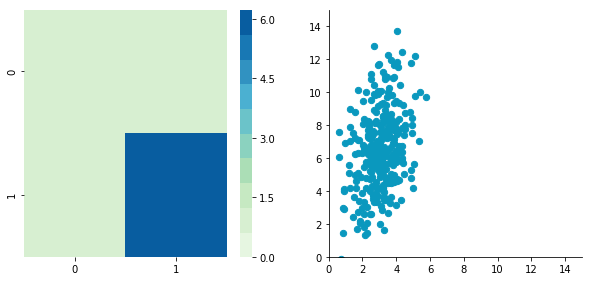
Correlated data
Now, let’s construct dependent data by specifying one column from the other one.
That’s great! ⚡️ We now have all the tools to see different preprocessing techniques.
2. Preprocessing
A. Mean normalization
Mean normalization is just removing the mean from each observation.
$ \boldsymbol
It will have the effect of centering the data around 0. We will create the function center() to do that:
B. Standardization
The standardization is used to put all features on the same scale. The way to do it is to divide each zero-centered dimension by its standard deviation.
Let’s create another dataset with a different scale to check that it is working.
C. Whitening
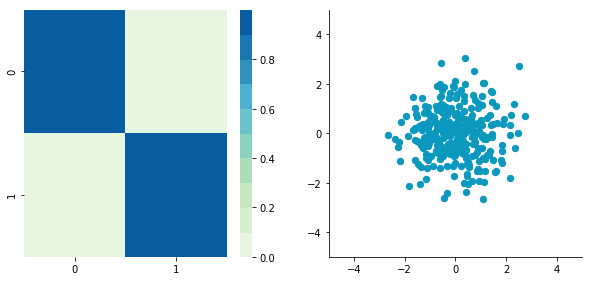
Whitening or sphering data means that we want to transform it in a way to have a covariance matrix that is the identity matrix (1 in the diagonal and 0 for the other cells; more details on the identity matrix). It is called whitening in reference to white noise.
We now have all the tools that we need to do it. It involves the following steps:
1. Zero-centering
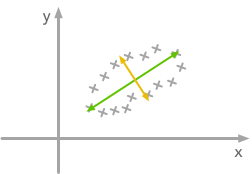
2. Decorrelate
At this point, we need to decorrelate our data. Intuitively, it means that we want to rotate the data until there is no correlation anymore. Look at the following cartoon to see what I mean:
The question is: how could we find the right rotation in order to get the uncorrelated data? Actually, it is exactly what the eigenvectors of the covariance matrix do: they indicate the direction where the spread of the data is at its maximum:
For more details about the eigendecomposition, see this post.
So we can decorrelate the data by projecting it on the eigenvectors basis. This will have the effect to apply the rotation needed and remove correlations between the dimensions. Here are the steps:
Let’s pack that into a function:
Nice! This is working 🎄
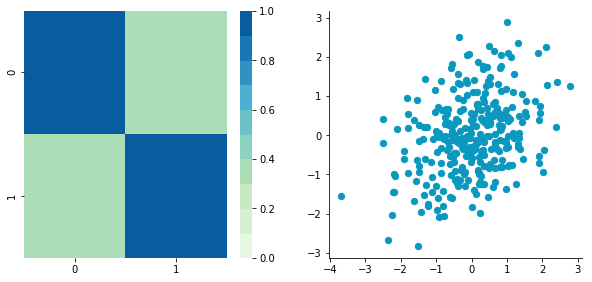
We can see that the correlation is not here anymore and that the covariance matrix (now a diagonal matrix) confirms that the covariance between the two dimensions is equal to 0.
3. Rescale the data
The next step is to scale the uncorrelated matrix in order to obtain a covariance matrix corresponding to the identity matrix (ones on the diagonal and zeros on the other cells). To do that we scale our decorrelated data by dividing each dimension by the square-root of its corresponding eigenvalue.
3. Image whitening
We will see how whitening can be applied to preprocess image dataset. To do so we will use the paper of Pal & Sudeep (2016) where they give some details about the process. This preprocessing technique is called Zero component analysis (ZCA).
Check out the paper, but here is the kind of result they got:

First thing first: we will load images from the CIFAR dataset. This dataset is available from Keras but you can also download it here.
The training set of the CIFAR10 dataset contains 50000 images. The shape of X_train is (50000, 32, 32, 3). Each image is 32px by 32px and each pixel contains 3 dimensions (R, G, B). Each value is the brightness of the corresponding color between 0 and 255.
We will start by selecting only a subset of the images, let’s say 1000:
The next step is to be able to see the images. The function imshow() from Matplotlib (doc) can be used to show images. It needs images with the shape ($M \times N \times 3$) so let’s create a function to reshape the images and be able to visualize them from the shape (1, 3072).
For instance, let’s plot one of the images we have loaded:
Remind that the formula to obtain the range [0, 1] is:
but here, the minimum value is 0, so this leads to:
Mean subtraction: per-pixel or per-image?
One way to do it is to take each image and remove the mean of this image from every pixel (Jarrett et al., 2009). The intuition behind this process is that it centers the pixels of each image around 0.
Another way to do it is to take each of the 3072 pixels that we have (32 by 32 pixels for R, G and B) for every image and subtract the mean of that pixel across all images. This is called per-pixel mean subtraction. This time, each pixel will be centered around 0 according to all images. When you will feed your network with the images, each pixel is considered as a different feature. With the per-pixel mean subtraction, we have centered each feature (pixel) around 0. This technique is commonly used (e.g Wan et al., 2013).
This gives us 3072 values which is the number of means: one per pixel. Let’s see the kind of values we have:
This is near 0.5 because we already have normalized to the range [0, 1]. However, we still need to remove the mean from each pixel:
Just to convince ourselves that it worked, we will compute the mean of the first pixel. Let’s hope that it is 0.
This is not exactly 0 but it is small enough that we can consider that it worked! 🌵
Now we want to calculate the covariance matrix of the zero-centered data. Like we have seen above, we can calculate it with the np.cov() function from Numpy.
To do so, we will tell this to Numpy with the parameter rowvar=False (see the doc): it will use the columns as variables (or features) and the rows as observations.
The covariance matrix should have a shape of 3072 by 3072 to represent the correlation between each pair of pixels (and there are 3072 pixels):
Now the magic part: we will calculate the singular values and vectors of the covariance matrix and use them to rotate our dataset. Have a look at my post on the singular value decomposition if you need more details!
In the paper, they used the following equation:
We will try to implement this equation. Let’s start by checking the dimensions of the SVD:
which corresponds to the shape of the initial dataset after transposition. Nice!
Let’s rescale the images:
Finally, we can look at the effect of whitening by comparing an image before and after whitening:
I hope that you found something interesting in this article!
You can fork the Jupyter notebook on Github here!
Что такое data whitening
Principal Components Analysis (PCA) is a dimensionality reduction algorithm that can be used to significantly speed up your unsupervised feature learning algorithm. More importantly, understanding PCA will enable us to later implement whitening, which is an important pre-processing step for many algorithms.
Suppose you are training your algorithm on images. Then the input will be somewhat redundant, because the values of adjacent pixels in an image are highly correlated. Concretely, suppose we are training on 16×16 grayscale image patches. Then \textstyle x \in \Re^ <256>are 256 dimensional vectors, with one feature \textstyle x_j corresponding to the intensity of each pixel. Because of the correlation between adjacent pixels, PCA will allow us to approximate the input with a much lower dimensional one, while incurring very little error.
Example and Mathematical Background
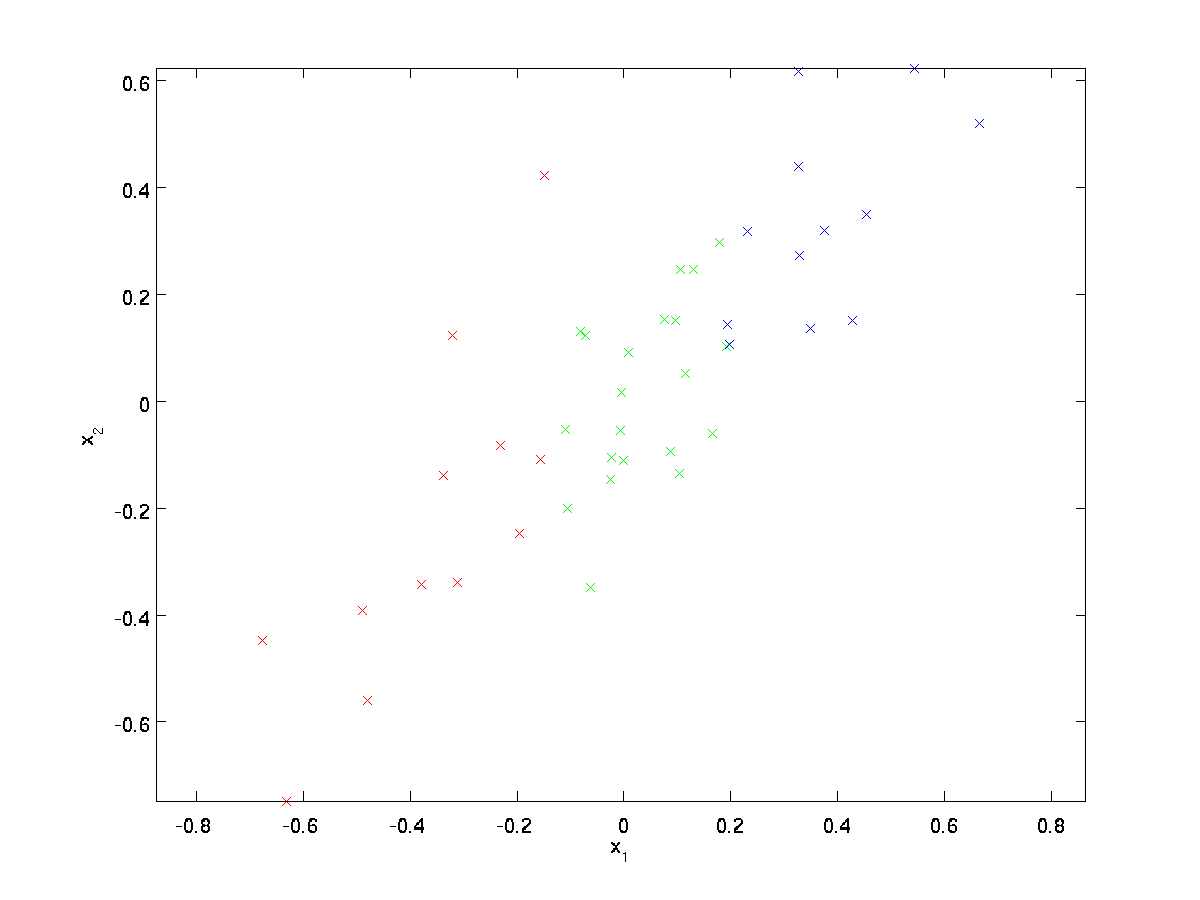
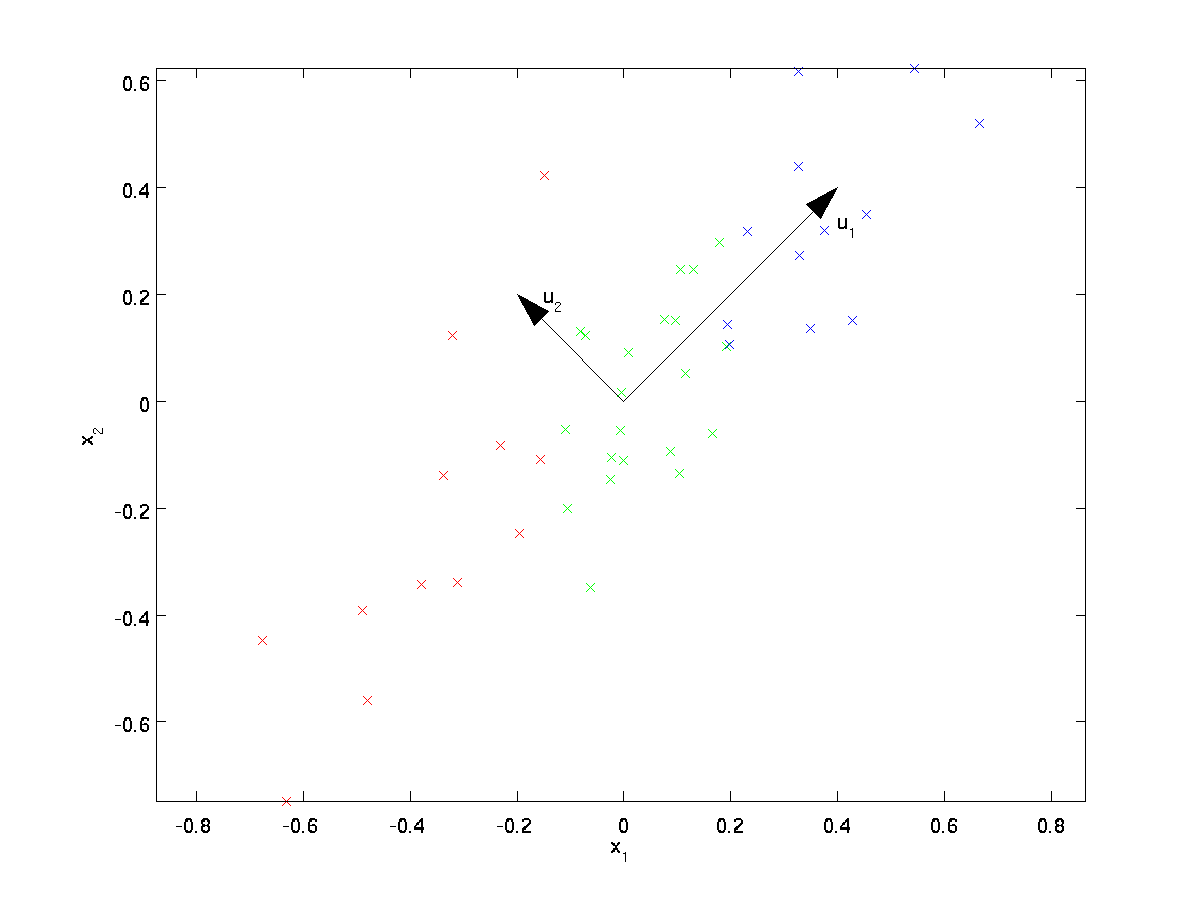
This data has already been pre-processed so that each of the features \textstyle x_1 and \textstyle x_2 have about the same mean (zero) and variance.
For the purpose of illustration, we have also colored each of the points one of three colors, depending on their \textstyle x_1 value; these colors are not used by the algorithm, and are for illustration only.
PCA will find a lower-dimensional subspace onto which to project our data.
From visually examining the data, it appears that \textstyle u_1 is the principal direction of variation of the data, and \textstyle u_2 the secondary direction of variation:
Note: If you are interested in seeing a more formal mathematical derivation/justification of this result, see the CS229 (Machine Learning) lecture notes on PCA (link at bottom of this page). You won’t need to do so to follow along this course, however.
\begin
Here, \textstyle u_1 is the principal eigenvector (corresponding to the largest eigenvalue), \textstyle u_2 is the second eigenvector, and so on. Also, let \textstyle \lambda_1, \lambda_2, \ldots, \lambda_n be the corresponding eigenvalues.
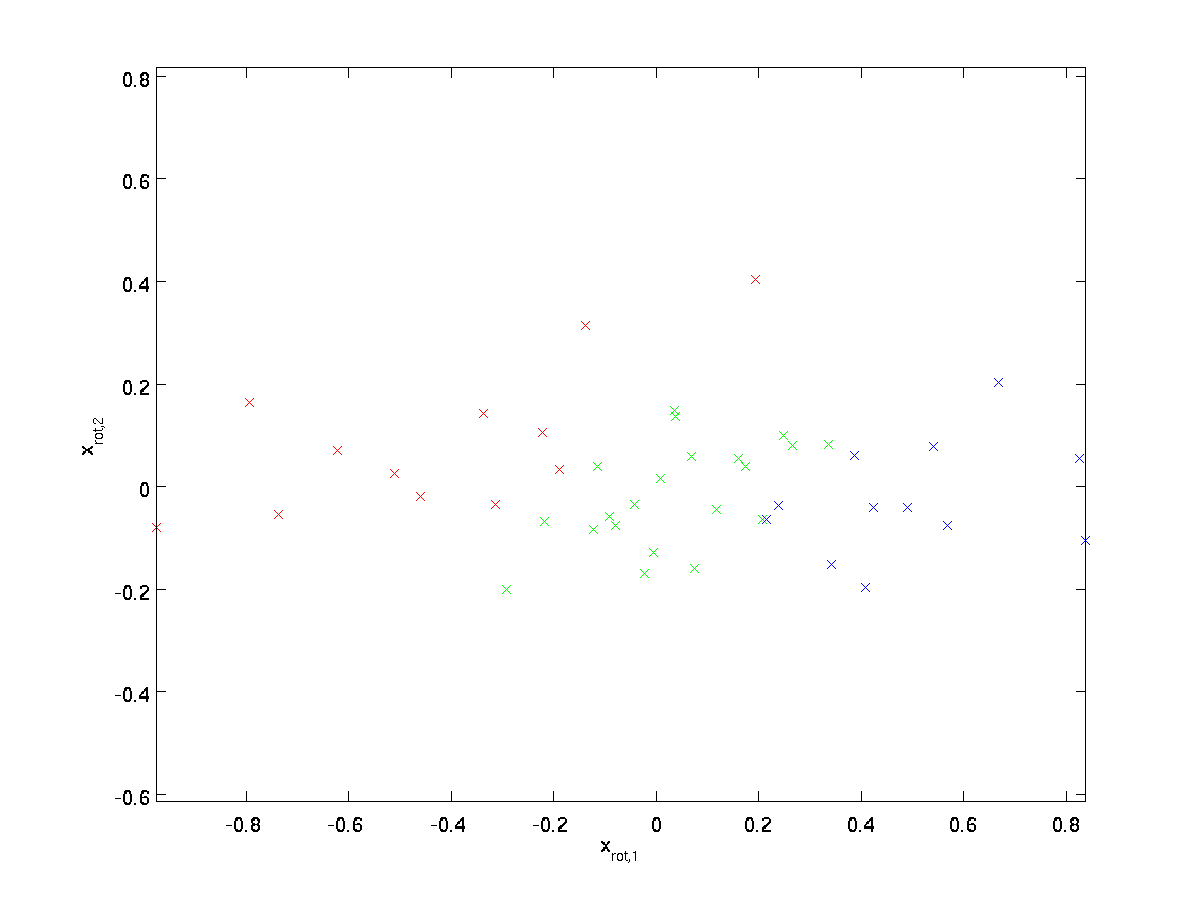
Rotating the Data
\begin
Reducing the Data Dimension
We see that the principal direction of variation of the data is the first dimension \textstyle x_ <\rm rot,1>of this rotated data. Thus, if we want to reduce this data to one dimension, we can set
More generally, if \textstyle x \in \Re^n and we want to reduce it to a \textstyle k dimensional representation \textstyle \tilde
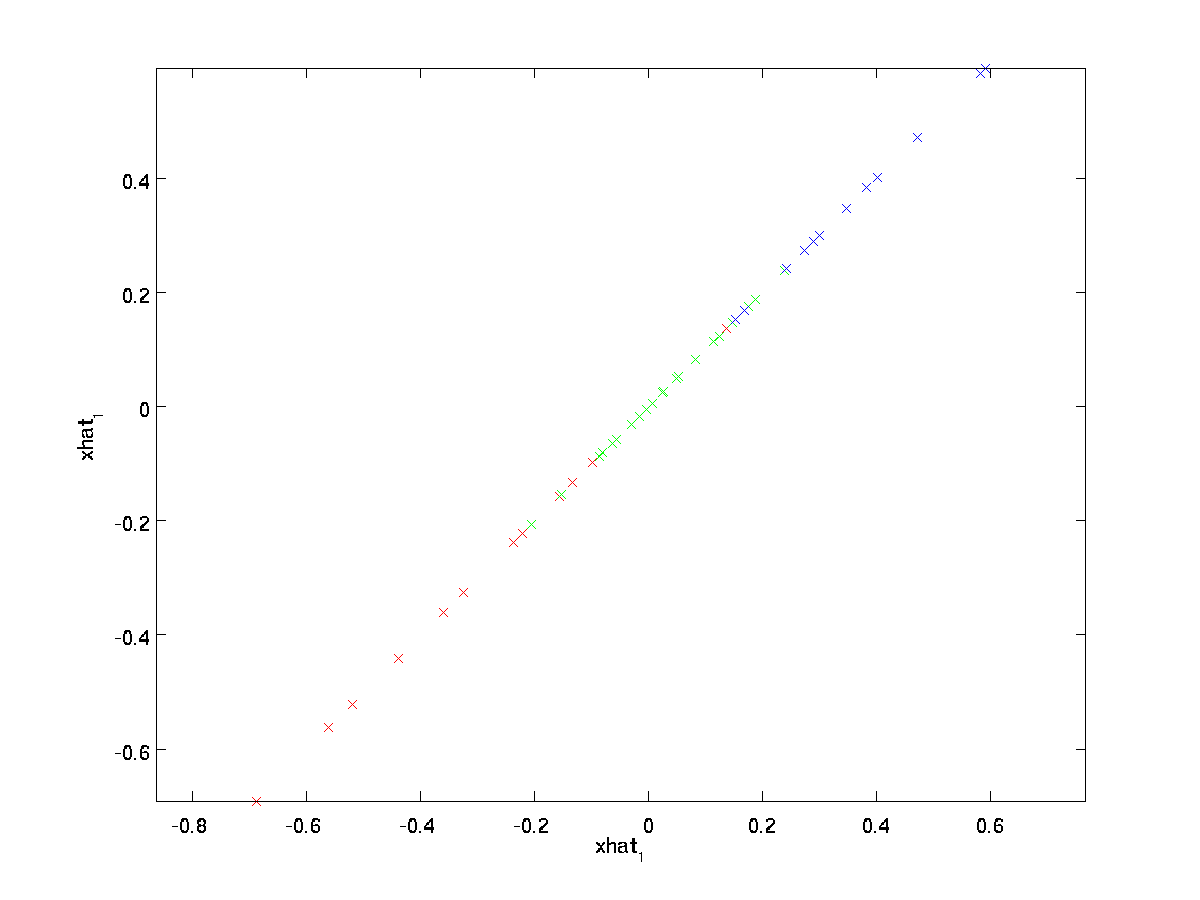
Another way of explaining PCA is that \textstyle x_ <\rm rot>is an \textstyle n dimensional vector, where the first few components are likely to be large (e.g., in our example, we saw that \textstyle x_<\rm rot,1>^ <(i)>= u_1^Tx^ <(i)>takes reasonably large values for most examples \textstyle i ), and the later components are likely to be small (e.g., in our example, \textstyle x_<\rm rot,2>^ <(i)>= u_2^Tx^ <(i)>was more likely to be small). What PCA does it it drops the the later (smaller) components of \textstyle x_ <\rm rot>, and just approximates them with 0’s. Concretely, our definition of \textstyle \tilde
In our example, this gives us the following plot of \textstyle \tilde
This also explains why we wanted to express our data in the \textstyle u_1, u_2, \ldots, u_n basis: Deciding which components to keep becomes just keeping the top \textstyle k components. When we do this, we also say that we are “retaining the top \textstyle k PCA (or principal) components.”
Recovering an Approximation of the Data
\begin
We are thus using a 1 dimensional approximation to the original dataset.
Number of components to retain
In the case of images, one common heuristic is to choose \textstyle k so as to retain 99% of the variance. In other words, we pick the smallest value of \textstyle k that satisfies
\begin
Depending on the application, if you are willing to incur some additional error, values in the 90-98% range are also sometimes used. When you describe to others how you applied PCA, saying that you chose \textstyle k to retain 95% of the variance will also be a much more easily interpretable description than saying that you retained 120 (or whatever other number of) components.
PCA on Images
Note: Usually we use images of outdoor scenes with grass, trees, etc., and cut out small (say 16×16) image patches randomly from these to train the algorithm. But in practice most feature learning algorithms are extremely robust to the exact type of image it is trained on, so most images taken with a normal camera, so long as they aren’t excessively blurry or have strange artifacts, should work.
When training on natural images, it makes little sense to estimate a separate mean and variance for each pixel, because the statistics in one part of the image should (theoretically) be the same as any other.
This property of images is called ”‘stationarity.”’
In detail, in order for PCA to work well, informally we require that (i) The features have approximately zero mean, and (ii) The different features have similar variances to each other. With natural images, (ii) is already satisfied even without variance normalization, and so we won’t perform any variance normalization.
(If you are training on audio data—say, on spectrograms—or on text data—say, bag-of-word vectors—we will usually not perform variance normalization either.)
In fact, PCA is invariant to the scaling of the data, and will return the same eigenvectors regardless of the scaling of the input. More formally, if you multiply each feature vector \textstyle x by some positive number (thus scaling every feature in every training example by the same number), PCA’s output eigenvectors will not change.
So, we won’t use variance normalization. The only normalization we need to perform then is mean normalization, to ensure that the features have a mean around zero. Depending on the application, very often we are not interested in how bright the overall input image is. For example, in object recognition tasks, the overall brightness of the image doesn’t affect what objects there are in the image. More formally, we are not interested in the mean intensity value of an image patch; thus, we can subtract out this value, as a form of mean normalization.
Concretely, if \textstyle x^ <(i)>\in \Re^
for all \textstyle j
If you are training your algorithm on images other than natural images (for example, images of handwritten characters, or images of single isolated objects centered against a white background), other types of normalization might be worth considering, and the best choice may be application dependent. But when training on natural images, using the per-image mean normalization method as given in the equations above would be a reasonable default.
Whitening
We have used PCA to reduce the dimension of the data. There is a closely related preprocessing step called whitening (or, in some other literatures, sphering) which is needed for some algorithms. If we are training on images, the raw input is redundant, since adjacent pixel values are highly correlated. The goal of whitening is to make the input less redundant; more formally, our desiderata are that our learning algorithms sees a training input where (i) the features are less correlated with each other, and (ii) the features all have the same variance.
2D example
We will first describe whitening using our previous 2D example. We will then describe how this can be combined with smoothing, and finally how to combine this with PCA.
How can we make our input features uncorrelated with each other? We had already done this when computing \textstyle x_<\rm rot>^ <(i)>= U^Tx^ <(i)>.
Repeating our previous figure, our plot for \textstyle x_ <\rm rot>was:
The covariance matrix of this data is given by:
\begin
(Note: Technically, many of the statements in this section about the “covariance” will be true only if the data has zero mean. In the rest of this section, we will take this assumption as implicit in our statements. However, even if the data’s mean isn’t exactly zero, the intuitions we’re presenting here still hold true, and so this isn’t something that you should worry about.)
To make each of our input features have unit variance, we can simply rescale each feature \textstyle x_ <\rm rot,i>by \textstyle 1/\sqrt <\lambda_i>. Concretely, we define our whitened data \textstyle x_ <\rm PCAwhite>\in \Re^n as follows:
Plotting \textstyle x_ <\rm PCAwhite>, we get:
Whitening combined with dimensionality reduction. If you want to have data that is whitened and which is lower dimensional than the original input, you can also optionally keep only the top \textstyle k components of \textstyle x_ <\rm PCAwhite>. When we combine PCA whitening with regularization (described later), the last few components of \textstyle x_ <\rm PCAwhite>will be nearly zero anyway, and thus can safely be dropped.
ZCA Whitening
Finally, it turns out that this way of getting the data to have covariance identity \textstyle I isn’t unique. Concretely, if \textstyle R is any orthogonal matrix, so that it satisfies \textstyle RR^T = R^TR = I (less formally, if \textstyle R is a rotation/reflection matrix), then \textstyle R \,x_ <\rm PCAwhite>will also have identity covariance.
Plotting \textstyle x_ <\rm ZCAwhite>, we get:
When using ZCA whitening (unlike PCA whitening), we usually keep all \textstyle n dimensions of the data, and do not try to reduce its dimension.
Regularizaton
When implementing PCA whitening or ZCA whitening in practice, sometimes some of the eigenvalues \textstyle \lambda_i will be numerically close to 0, and thus the scaling step where we divide by \sqrt <\lambda_i>would involve dividing by a value close to zero; this may cause the data to blow up (take on large values) or otherwise be numerically unstable. In practice, we therefore implement this scaling step using a small amount of regularization, and add a small constant \textstyle \epsilon to the eigenvalues before taking their square root and inverse:
For the case of images, adding \textstyle \epsilon here also has the effect of slightly smoothing (or low-pass filtering) the input image. This also has a desirable effect of removing aliasing artifacts caused by the way pixels are laid out in an image, and can improve the features learned (details are beyond the scope of these notes).
ZCA whitening is a form of pre-processing of the data that maps it from \textstyle x to \textstyle x_ <\rm ZCAwhite>. It turns out that this is also a rough model of how the biological eye (the retina) processes images. Specifically, as your eye perceives images, most adjacent “pixels” in your eye will perceive very similar values, since adjacent parts of an image tend to be highly correlated in intensity. It is thus wasteful for your eye to have to transmit every pixel separately (via your optic nerve) to your brain. Instead, your retina performs a decorrelation operation (this is done via retinal neurons that compute a function called “on center, off surround/off center, on surround”) which is similar to that performed by ZCA. This results in a less redundant representation of the input image, which is then transmitted to your brain.
Implementing PCA Whitening
In this section, we summarize the PCA, PCA whitening and ZCA whitening algorithms, and also describe how you can implement them using efficient linear algebra libraries.
First, we need to ensure that the data has (approximately) zero-mean. For natural images, we achieve this (approximately) by subtracting the mean value of each image patch.
We achieve this by computing the mean for each patch and subtracting it for each patch. In Matlab, we can do this by using
(Note: The svd function actually computes the singular vectors and singular values of a matrix, which for the special case of a symmetric positive semi-definite matrix—which is all that we’re concerned with here—is equal to its eigenvectors and eigenvalues. A full discussion of singular vectors vs. eigenvectors is beyond the scope of these notes.)
Finally, you can compute \textstyle x_ <\rm rot>and \textstyle \tilde
To compute the PCA whitened data \textstyle x_ <\rm PCAwhite>, use
Finally, you can also compute the ZCA whitened data \textstyle x_ <\rm ZCAwhite>as: